This blog supports the CH795 Special Topics in Chemistry courses taught by Dr. Gavin Williams and Dr. Alex Deiters at North Carolina State University. Please include an illustrative figure when you post a blog entry.
Tuesday, August 23, 2011
The rise, fall and reinvention of combinatorial chemistry
We talked about small molecule libraries, library assembly, and library diversity in class today. This is a short highlight by Tom Kodadek (Scripps FL) in which he summarizes the most recent developments. Give it a quick read, since some of the discussed technologies will be discussed in class (e.g., DNA-templated synthesis and DNA-encoded libraries).
Subscribe to:
Post Comments (Atom)
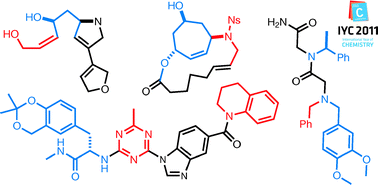
As the author pointed out Diversity-Oriented Synthesis is a great improvement for combinatorial Chemistry. Since it yields to much more diverse compounds, the chances to find 'hits' are higher. On the other hand, these compounds are often really challenging to synthesize, they have lots of stereocenters. Whereas when performing Structure-Activity Relationship, it is preferable to work with readily accessible compounds. I guess DOS is a great tool but it is not that effective on a medicinal chemistry level.
ReplyDeleteThe author mentions the small molecules attached to beads. Are the chemical reactions performed with various beads, each containing a different type of small molecule, acting as the 'starting material'? It would be really efficient to do one chemical reaction, but with so many beads/skeletons that you are actually creating thousands of different compounds. Is this why the resulting library can be considered un-biased, because the chemist isn't actively trying to arrive at a particular structure? ...Or maybe I'm understanding this wrong and the beads were just used for screening?
ReplyDeleteAlso, the phage library blew my mind.
What you are thinking of is actually a very old concept in combinatorial chemistry: it's called 'split-and-pool synthesis'. The Schreiber lab has a lovely animation explaining it: http://www.broadinstitute.org/chembio/lab_schreiber/anims/animations/smdbSplitPool.php
ReplyDeletePS: Please use real names on this blog!
The library is called "unbiased" because the compounds are not designed toward some specific target. The goal is to get the maximum of diversity among compounds that have the same core structure to increase the chances of finding hits when these compounds are screened toward different proteins.
ReplyDelete